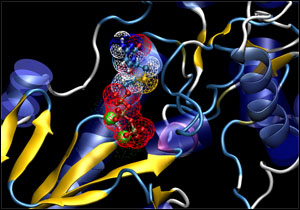
Lamivudine bound in HBV polymerase
Drug Development
The SeqHepB database, underpinned by an extensive patent portfolio of HBV mutations, can be used by major pharmaceutical companies for cross-resistance testing of their newer generation HBV drugs currently under development.
These new anti-HBV agents, presently in clinical trials in the USA, require monitoring of HBV drug resistance, especially with the revised licensing regulations of new antiviral agents as demanded by FDA. SeqHepB database can be utilized to analyse the virological content of the phase I, II or III clinical trial data.
SeqHepB can assist in the development of the next generation of drugs targeting HBV. Rational drug design bases the invention of the new drug on the structure of its protein target.
This approach has been enhanced recently through use of combinatorial chemistry and high throughput screening and has been highly successful in the field of HIV with the protease inhibitor drugs.
The SeqHepB viral genomic database can be linked to a computer-based three-dimensional molecular model of the HBV polymerase protein, which is the target of the current antiviral agents. Linking the computer model to the genotype database of HBV drug resistant mutants enables drug developers to potentially identify potential chemical alterations to the antiviral agent needed to generate a novel and more active compound.
This facilitates analysis of patients' HBV polymerase and potential mechanisms of antiviral resistance and this approach may provide opportunities for improved novel drug design.
All material copyright Evivar Medical Pty. Ltd. All rights reserved